An HDF5 primer¶
The currently recommended file format for NeXus is HDF5. Hence, understanding the basic concepts behind HDF5 is absolutely mandatory to work with NeXus in a productive way.
HDF5 differs from the commonly used file formats in two fundamental ways
- It is a binary format.
- The data is organized as a tree rather than as tables.
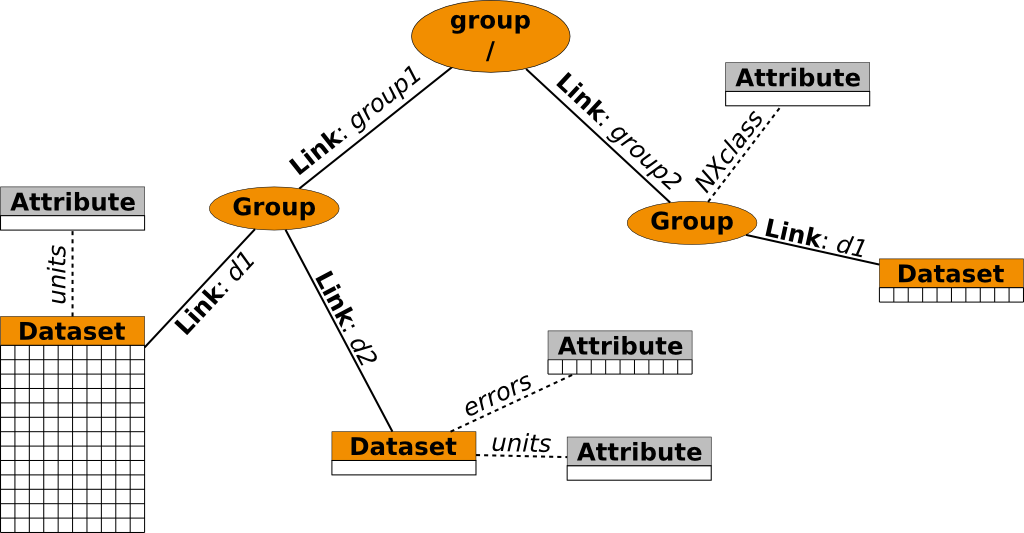
An HDF5 data tree is represented using the following basic objects
- groups which are the nodes of the tree
- datasets which are the data storing leaves
- links which connect nodes and leaves
- attributes which are, with some limitations, similar to datasets and can be attached to groups and datasets to store additional meta-data.
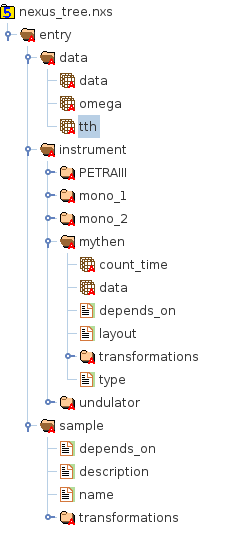
Though this sounds rather complex, in practice, an HDF5 tree looks quite similar to a file system tree.
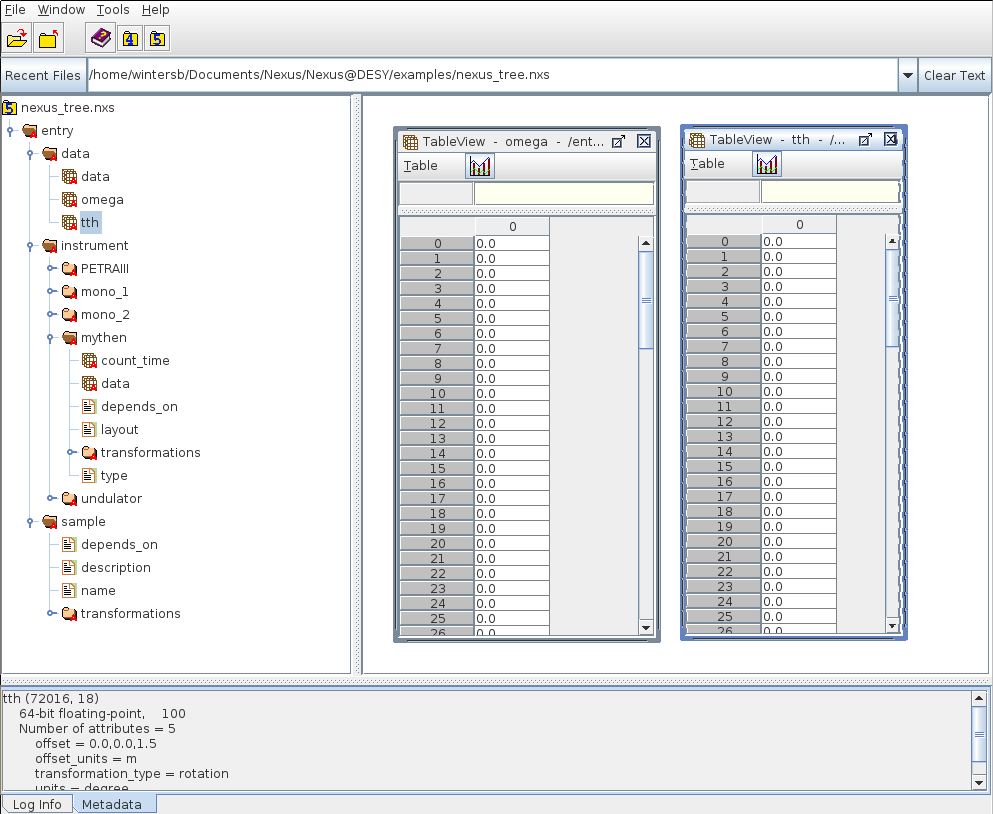
HDFview showing data stored in an HDF5 file. The left panel shows the data tree which looks quite similar to a filesystem tree.
In this picture the groups represent directories, datasets are the files in each directory and links can be hard- or symbolic-links as they are used on common filesystems. There is no equivalent the HDF5 attributes in this picture but this is not a serious limitation.
Paths and links¶
HDF5 uses a path syntax similar to Unix file systems to address objects within a file.
To access the data field in the mythen group the following path can be used
/entry/instrument/mythen/data
The connections between the objects resembling an HDF5 file are made by links. HDF5 has 3 kinds of links
- hard links - which is used when a new object is created
- soft links - like symbolic links on a filesystem they can be used to refer to an existing object at a different location
- and external links - which can be used to reference objects in a different file.
For users in particular the latter two are of importance. Soft links can be used to avoid data duplication. In the above example the datasets below /entry/data for instance are soft-links to datasets stored somewhere else in the file. All links in HDF5 are completely transparent to the user. This means that the procedure to access a dataset does not depend on whether or not the access is made via a hard-, a soft-, or an external-link.
However there are some pitfalls users should be aware about. The most important one is that like on a filesystem, soft- and external-links can be created whether or not the target (the object the link refers to) exists. If it does not, trying to access the object via the link will cause a failure. Aside from this rather obvious problem there is an additional one which particularly concerns external links.
It is strongly recommended to use relative paths (relative to the location of the file where the link should be created) when creating external links. This ensures that the link can be resolved even when the directory structure the files reside in is copied to a different location. However, there is a small inconvenience. Consider that the path to the target file is relative to the location of the file where the link is created. If we try to access the data via the external link the program trying to access the data should have its current directory set to the location of the file with the link. Otherwise the resolution of the external link will fail.
Compression of individual datasets¶
To reduce the amount of disk space occupied by a data file on might think about using compression to reduce its size. However, compressing the entire file has several disadvantages
- not all data within the file are compressable with reasonable data reduction
- compressing the entire file would require quite some work to access small amounts of meta-data stored within it.
- different data within the file cannot be compressed with different algorithms.
To overcome these issues HDF5 provides the functionality to apply compression to individual datasets rather than to the entire file. Consider for instance a file with data from an experiment with a large 2D detector. The detector images are stored in a 3D dataset as a single block. However, they occupy 95% of the amount of disk space occupied by the file. Everything else is mall meta-data. With HDF5 it is possible to apply compression only to the dataset with the 3D data. Thus, retrieving meta-data is as fast as before but the ammount of disk space required is significantly reduced. In particular with low-noise detectors this makes a lot of sense.
It is also possible to use different compression algorithms for different datasets.